Abstract
Background: Kidney failure occurs in 5-13% of individuals with sickle cell disease (SCD), and is associated with early mortality. We have shown previously that genetic variation in apolipoprotein L1 (APOL1) is associated with increased risk for developing SCD nephropathy (Ashley-Koch et al, 2011). Two APOL1 alleles (G1 and G2) have been identified as risk factors for SCD nephropathy, as well as chronic kidney disease (CKD) in African Americans without SCD (Limou et al, 2014). Both risk alleles are highly prevalent in individuals of African descent. Despite the strong association of G1 and G2 with kidney dysfunction, the mechanisms by which these variants contribute to increased CKD risk remain poorly understood. However, the observation of podocyte effacement in CKD patients suggests that podocytes and/or endothelial cells are likely implicated in pathogenesis. We established zebrafish models of these human risk alleles previously for the purpose of better understanding the underlying pathology contributing to SCD nephropathy (Anderson et al, 2015). Here, we have utilized these models to explore further the cell-type specific transcriptomic perturbations contributing to nephropathy.
Methods: To investigate the effects of the APOL1 G2 risk variant on cell-type specific transcriptomes, we injected human APOL1 mRNA into embryos from 3 different zebrafish lines expressing cell-type specific reporters to permit the isolation of podocytes (pod:NTR-mCherry), endothelial cells (fli1:egfp), or peripheral neurons (hb9:gfp; control). Three populations from each line were utilized: (1) uninjected controls; (2) embryos injected with canonical human APOL1 (G0); and (3) embryos injected with human APOL1 (G2) mRNA. Larvae were dissociated at 4 days post-fertilization (4 dpf), and fluorescent cell types were isolated via fluorescence activated cell-sorting (FACS). Messenger RNA was extracted from sorted cell populations (n=3 FACS sorted cell populations per condition) and subsequently amplified to obtain a sufficient amount of RNA for sequencing. The amplified RNA was prepared into cDNA libraries, and sequenced on an Illumina HiSeq 2500. We performed pairwise comparisons between uninjected and G0 injected larvae in each cell type to ascertain the effects of canonical human APOL1 on the zebrafish kidney transcriptome. Additionally, to measure the effects of APOL1 G2 expression on the transcriptome, we compared APOL1 G0 and APOL1 G2 samples. Transcripts were considered to be differentially expressed if they achieved an FDR-adjusted q-value of less than 0.05. To understand better the biological processes altered by injection condition, we conducted gene ontology enrichment analyses (GOEA) on sets of differentially expressed genes.
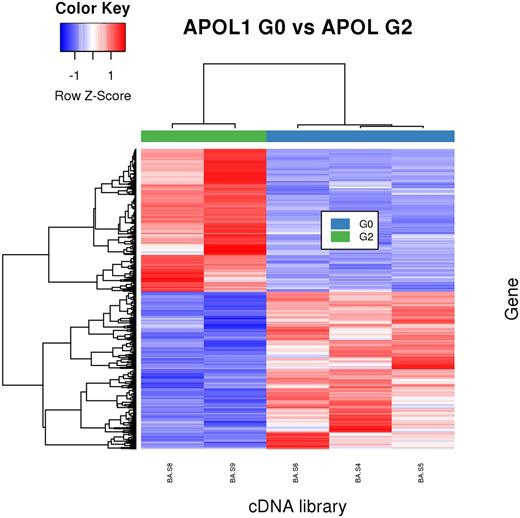
Results: Our analysis of uninjected and APOL1 G0 libraries identified 38, 154, and 204 differentially expressed (DE) genes in endothelial cells, podocytes, and motor neurons, respectively. No significant enrichment of gene ontology terms was detected, suggesting that canonical G0 APOL1 did not have a substantial impact on the zebrafish transcriptome. In contrast, the comparison of APOL1 G0 and APOL1 G2 samples found 95, 7400 (Figure below), and 652 DE genes in endothelial cells, podocytes, and motor neurons, respectively. GOEA of the podocyte DE gene list showed enrichment for several autophagy-associated terms, including "Lysosome" and "Phagosome". Close inspection of these autophagy-associated genes revealed that the majority are up-regulated in G2 relative to G0 (68/75 genes for "Lysosome" and 52/78 genes for "Phagosome"). Thus, the G2 allele appears to induce transcription of genes involved in autophagy in podocytes.
Summary: Taken together, these data suggest that podocytes are the predominant cell type affected by the expression of APOL1 G2. The comparison of podocytes injected with APOL1 G0 or APOL1 G2 yielded an order of magnitude more DE transcripts than any other comparison. The GOEA results of the podocyte comparison are consistent with a recent study of transgenic mice expressing the G1 or G2 APOL1 high risk variants, which showed alterations in endocytic vesicle content and autophagic flux (Beckerman et al 2017). Ongoing work includes molecular analysis to characterize the effects of the APOL1 G1 allele. This work will continue to shed light on the molecular pathology of SCD nephropathy and provide insight into new therapeutic strategies.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal